Agricultural and Biological Research
RNI # 24/103/2012-R1
Research Article - (2022) Volume 38, Issue 2
The Bacterial leaf blight of rice caused by Xanthomonas oryzae pv. oryzae is one of the most devastating pathogenic diseases responsible for extensive crop loss across different rice growing regions of the world. Presently, new X. oryzae strains have emerged which has managed to overcome the resistance conferred by major genes like xa5 and xa13. Under such circumstances the current paper evaluates rice genotypes belonging to diverse origins, viz. high yielding varieties, indigenous landraces and IR24 near isogenic lines, in terms of their resistance against three widely available strains of X. oryzae in India vis-à-vis West Bengal. The genotypes were also screened using molecular markers which are tightly linked with four major resistance conferring genes, namely Xa4, xa5, xa13 and Xa21. The overall analysis revealed that the three gene combination i.e. xa5+xa13+Xa21 was essential for maintaining stability in resistance against all the three pathogenic strains used for inoculation in the current study. A deviation from the above mentioned gene combination resulted in variable degree of pathogenesis, depending upon the Xoo strain used for inoculation. The investigation also identified the presence of dominant Xa4 gene among many of the popular cultivars in West Bengal, as well as in indigenous landraces like Kalabhat and Chamarmani; which showed strong resistance against the local Xoo strain. However, the genotypes failed to show any significant resistance against newly introduced strains indicating that the popular cultivars in West Bengal are still vulnerable towards new pathogenic strains of Xanthomonas oryzae. Thus, introgression of multiple resistance genes into popular cultivars in the region is necessary for ensuring durable resistance against the disease.
Mineral nutrients; Edible plants; Dry climate; Fertilizer
Rice (Oryza sativa L.) is one of the most important staple foods consumed by more than half of the world’s population and occupies almost one-fifth of the total land area covered under cereals [1,2]. Bacterial leaf blight in its most aggressive form can cause up to 50% yield loss across different rice growing areas in India [3]. Mew indicated that the disease is most vigorous under a rain- fed low land ecosystem, which is quite common across different rice growing areas in West Bengal as well as in India [4]. Mishra isolated 1024 Xoo strains from 20 states in India and identified eleven distinct pathotypes which were widely distributed across all the states [5]. Currently, 40 BLB resistance genes have been indentified, conferring various degrees of resistance against the pathogen [6]. Among the genes identified, four major resistance conferring genes, namely Xa4, xa5, xa13 and Xa21 have been primarily utilized by rice breeders for ensuring resistance against multiple strains of X. oryzae. The gene combination Xa4+xa5+xa13+Xa21 have been introgressed into indigenous rice variety Jalamaghna in the works of Pradhan [7]. In recent times reports have emerged from India and Thailand regarding X. oryzae strains which have overcome the resistance conferred by xa5 and xa13 genes [8]. Rice being the staple diet in West Bengal along with large parts of India, monitoring of BLB pathogenesis among popular rice cultivars can help in predicting the extent of damage expected for these cultivars, especially in the advent of a X. oryzae epidemic. The present investigation analyses the variability in pathogenesis recorded for 20 diverse rice genotypes (possessing varying degrees of resistance genes) in response to three pathogenic strains of X. oryzae which are widely observed in different rice growing regions of India.
Twenty genotypes comprising of four near isogenic lines (NILs) of IR24 and sixteen popular cultivars were screened under greenhouse conditions for detecting their disease response against three strains of Xanthomonas oryzae pv. oryzae. The experiment was conducted for two successive years of 2018 and 2019 at the University of Calcutta, India (Table 1).
| Sl.no. | Name of germplasms | Origin |
|---|---|---|
| 1 | IR24(SC) | IRRI |
| 2 | IR36 | IRRI |
| 3 | Carolina gold | Indonesia |
| 4 | IR64 | IRRI |
| 5 | Jaldi 13 | NRRI, Cuttack |
| 6 | IRBB21(control for Xa21) | IRRI |
| 7 | IRRI146 | IRRI |
| 8 | BJ1 | Bangladesh |
| 9 | IRBB60 (RCI) | IRRI |
| 10 | IR29 | IRRI |
| 11 | IRBB59 (RCII) | IRRI(IR24 NIL) |
| 12 | IRBB13(control for xa13) | IRRI(IR24 NIL) |
| 13 | IR72 | IRRI |
| 14 | SWARNA | APRRI and RARS, Maruteru |
| 15 | VANDANA | CRRI, Cuttack |
| 16 | Kalabhat | Indian land race |
| 17 | Kalonunia | Indian land race |
| 18 | Chamarmani | Indian land race |
| 19 | Kerala Sundari | Indian land race |
| 20 | Dudheswar | Indian land race |
Note: SC= susceptible check, RC= Resistant check
Table 1 List of genotypes screened under greenhouse conditions for their response against three strains of Xanthomonas oryzae
The IR24 near isogenic lines IRBB60 and IRBB59 were designated as resistant checks, carrying the Xa4+xa5+xa13+Xa21 and xa5+xa13+Xa21 resistance gene combinations respectively. The NILs IRBB13 and IRBB21 were designated as controls for the gene xa13 and Xa21 respectively. The genotype IR24 was designated as the susceptible check.
Green house screening
The twenty genotypes evaluated in the current experiment were germinated in sterilized petri-plates containing moist filter papers using ultrapure Mili-Q water. Following germination and seedling emergence, the seedlings were placed in pots of 20 cm diameter. The pot soil was sterilized by autoclaving at 121oC for 20 minutes. Twenty one days old seedlings were transferred to larger pots of 40 cm diameter. 1.0 g of N.P.K at 15:15:15 dose was added along with 0.2 g of 46% Urea at 15 days from transplanting into large pots, as suggested in the works of Banito [9]. Plants were watered at alternate days. The greenhouse temperature was set at a range of 28 to 30oC. The relative humidity was maintained at 90 to 95%.
Maintaining of Xoo strains
The purified isolates of X. oryzae were maintained in 500 μl of 20% glycerol at - 20°C. Three strains of X. oryzae which are widely available in India were used for the experiment. Two strains, viz. IX021 and IX027 were collected from IIRR (Indian Institute of Rice Research). The third strain was a local isolate collected from Rice Research Station, Chinsurah, and West Bengal, India. The local isolate was designated as CUXo1.
Inoculation of genotypes with the three pathogenic strains
The bacterial suspension was adjusted to a concentration of 108 cfu ml-1 and the host plants were inoculated using the clipping method suggested by Kauffman [10]. The method involved clipping the leaf tip using scissors dipped in bacterial suspension at a height of 3 cm from the tip of a healthy leaf. The inoculation was carried out during active tillering stage of the host plants.
Collection of data associated with lesion length
Infected leaves from three replicates of each genotype inoculated separately by the three strains were collected and lesion length was measured at fourteen days after inoculation. Based on the lesion length recorded, the genotypes were classified as per IRRI (Table 2).
| Lesion length (cm) | Disease reaction |
|---|---|
| 0-5 | Resistance (R) |
| >5-10 | Moderately Resistant (MR) |
| >10-15 | Moderately Susceptible (MS) |
| >15 | Susceptible (S) |
Table 2 Scoring of genotypes for disease response
Screening of genotypes using molecular markers tightly linked with four BLB resistance genes Xa4, xa5, xa13 and Xa21
The marker assisted screening was performed using molecular markers which are tightly linked with four major resistance conferring genes, viz. Xa4, xa5, xa13 and Xa21 (Table 3). Plant DNA was extracted from leaf samples of 20 genotypes using DNeasy Plant Mini Kit (Maker: Qiagen). The polymerase chain reaction was conducted in a reaction solution of 25 µl containing template DNA, primers, dNTPs, reaction buffer, MgCl2 and Taq Polymerase. The PCR amplification was performed using an Eppendorf thermo-cycler. The PCR cycle profile involved 4 minutes of denaturation at 94°C followed by 35 cycles of 30 seconds denaturation at 94°C. Then a 60 seconds annealing was allowed at 55°C followed by 60 seconds extension at 72°C. Final extension was allowed at 72°C for 5 minutes. The PCR products were resolved using 1.5% Agarose gel for the marker pTA248. 2% Agarose gel was used for the rest of the markers. A 50 to 1000 bp long DNA ladder was used for analyzing the size of amplified products. Bands were visualized under UV light using the gel documentation system. Resistance and susceptible alleles were observed and recorded for further study.
| Genes | chromosome | Primers | Sequences | Reference |
|---|---|---|---|---|
| Xa4 | 11 | RM224F | atcgatcgatcttcacgagg | Sun et al. (2003) |
| RM224R | tgctataaaaggcattcggg | |||
| xa5 | 5 | xa5s-F (Multiplex) | gtctggaatttgctcgcgttcg | Pradhan et al. (2015) |
| xa5s-R (Multiplex) | tggtaaagtagataccttatcaaactgga | |||
| xa5sr/R-F(Multiplex) | agctcgccattcaagttcttgag | |||
| xa5sr/R-R(Multiplex) | tgacttggttctccaaggctt | |||
| xa13 | 8 | Xa13 Prom-F | ggccatggctcagtgtttat | Sundaram et al. (2011) |
| Xa13 Prom-R | gagctccagctctccaaatg | |||
| Xa21 | 11 | pTA248 F | agacgcggaagggtggttcccgga | Pradhan et al. (2015) |
| pTA248 R | agaccggtaatcgaaagatgaaa |
Table 3 Markers used for identifying three resistant genes towards BLB
Statistical analysis
The pooled data was analyzed using Duncan's multiple range test (DMRT) using SPSS version 20.0 [11]. The pooled LSD at 5% significance level was calculated from pooled analysis of variance. Cluster analysis was performed on the basis of the allelic data recorded for the genotypes tested; using gene specific molecular markers. UPGMA (Unweighted Pair Group Method with Arithmetic Mean) method of clustering was followed and a dendrogram was constructed using Darwin version 6.0 [12].
Twenty diverse genotypes were screened for analyzing their response towards three invasive strains of bacteria Xanthomonas oryzae. The average leaf lesion lengths were calculated over two years for the genotypes, which were inoculated with three pathogenic BLB strains (Table 4). For the Xoo strain IX021, a formidable resistance was observed among the check varieties IRBB60 and IRBB59; which could not be matched by any other genotype except IRBB21. The similar response of the three genotypes was further confirmed by the Duncan's multiple range tests.
| Sl.no. | Genotypes | IX021 | Response | IX027 | Response | CUXo1 | Response |
|---|---|---|---|---|---|---|---|
| 1 | IR24 (SC) | 16ab | S | 17.16a | S | 15.8abc | S |
| 2 | IR36 | 15.33abc | S | 16.16ab | S | 15.93abc | S |
| 3 | Carolina gold | 6.83d | MR | 10.16cd | MS | 8.73e | MR |
| 4 | IR64 | 9d | MR | 17.26a | S | 16.26ab | S |
| 5 | JALDI 13 | 15.33abc | S | 14.2ab | MS | 16.33ab | S |
| 6 | IRBB21 | 3.16e | R | 8.5d | MR | 6.5f | MR |
| 7 | IRRI146 | 13.33bc | MS | 12.93bc | MS | 17.46a | S |
| 8 | BJ1 | 16.5ab | S | 3.33e | R | 15.06bc | S |
| 9 | IRBB60 (RC) | 1.33e | R | 4.16e | R | 0.46g | R |
| 10 | IR29 | 12.33abc | MS | 13.86ab | MS | 16.43ab | S |
| 11 | IRBB59 (RC) | 2.83e | R | 0.86e | R | 0.86g | R |
| 12 | IRBB13 | 8.83d | MR | 16.03ab | S | 12.33d | MS |
| 13 | IR72 | 17.5a | S | 14.7ab | MS | 13.9bc | MS |
| 14 | Swarna | 7.66d | MR | 14.26ab | MS | 15.96abc | S |
| 15 | Vandana | 16.66ab | S | 16.23ab | S | 15.03bc | S |
| 16 | Kalabhat | 7.20d | MR | 9.50d | MS | 4.10h | R |
| 17 | Kalonunia | 16.43ab | S | 17.63a | S | 18.43a | S |
| 18 | Chamarmani | 6.03d | MR | 8.83d | MS | 2.13h | R |
| 19 | Kerala Sundari | 17.23a | S | 16.03 ab | S | 17.13a | S |
| 20 | Dudheswar | 12.70abc | MS | 13.80ab | MS | 12.50d | MS |
| LSD at 5% probability | 3.511 | 3.814 | 1.519 |
Note: a, b, c, d, e, f = Pathogenicity Response
Table 4 Average Lesion length (cm) observed in twenty genotypes inoculated by three strains of X. oryzae across two years
On the other hand, a moderate degree of resistance was observable for the cultivars, namely Carolina Gold, IR64, IRBB13 and Swarna. Among the popular landraces indigenous to West Bengal, only Kalabhat and Chamarmani exhibited a moderate resistance against the strain IX021, whereas landraces like Kalonunia, Kerala Sundari and Dudheswar exhibited moderate to complete susceptibility against the strain. The results from the DMRT further confirm the similar pathogenicity observed among genotypes inoculated with IX021.
The disease development observed among the genotypes in response to the strain IX027 was also recorded. The investigation identified a single variety Bj1 which exhibited significant resistance against IX027, which was at par with the checks IRBB60 and IRBB59. The similar responses of the three genotypes were reconfirmed using DMRT.
Conversely, a moderate resistance was observable for IRBB21 in response to the pathogenic strain IX027. Rest of the genotypes exhibited susceptible or moderately susceptible reactions against the strain. The local landraces Kalabhat and Chamarmani failed to show significant resistance against the strain, instead exhibited moderate susceptibility. Other indigenous varieties like Kalonunia, Kerala Sundari and Dudheswar showed complete susceptibility against the strain.
Lastly, the genotypes were screened for analyzing their pathogenesis against the local isolate CUXo1. Other than the landraces Kalabhat and Chamarmani, no other genotype exhibited significant resistance at par with the (resistant) checks IRBB59 and IRBB60.
The genotypes Carolina Gold and IRBB21 on the other hand exhibited a moderate resistance against the strain. Other landraces analyzed in the current study, namely Kalonunia, Kerala Sundari and Dudheswar exhibited severe damage from BLB infection; indicating their susceptibility towards the strain. From the phenotypic observations it was evident that variability in terms of pathogenesis can be expected among the cultivars in response to the pathogenic strains, only exception being IRBB59 and IRBB60 which showed a stable resistance against the three strains. Variability in terms of pathogenesis of BLB has been earlier reported by Ogawa [13].
The phenotypic observations were corroborated at genotypic level using molecular markers which are tightly linked with four major resistance conferring genes, namely Xa4, xa5, xa13 and Xa21. The marker assisted screening of the genotypes elucidated the presence or absence of desirable alleles for the corresponding BLB resistance genes. The presence of the dominant allele for the gene Xa4 was detected using the marker RM224. The marker amplified a 160 bp resistance allele for the check variety IRBB60. On the other hand the susceptible check IR24 exhibited an allele size of 140 bp, when evaluated using the primer RM224. The 160 bp resistance allele similar to the check variety IRBB60 was also detected in the genotypes IR36, IR64, IR29 and JALDI 13 (Figure 1). Among the indigenous landraces the resistance allele for Xa4 was detected in genotypes, namely Kalabhat, Chamarmani and Dudheswar. The marker RM224 was successfully used for the detection of Xa4 gene among various rice genotypes in the works of Singh [14].
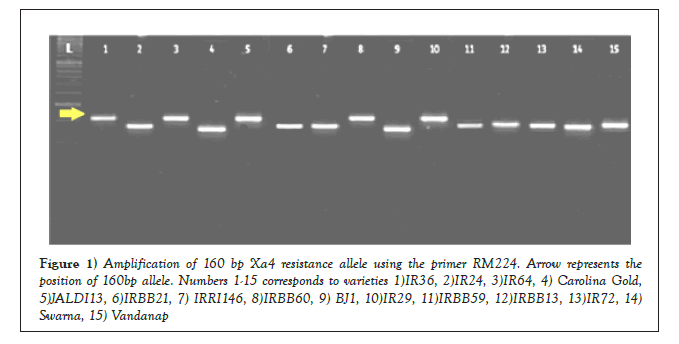
Figure 1: Amplification of 160 bp Xa4 resistance allele using the primer RM224. Arrow represents the position of 160bp allele. Numbers 1-15 corresponds to varieties 1)IR36, 2)IR24, 3)IR64, 4) Carolina Gold, 5)JALDI13, 6)IRBB21, 7) IRRI146, 8)IRBB60, 9) BJ1, 10)IR29, 11)IRBB59, 12)IRBB13, 13)IR72, 14) Swarna, 15) Vandanap
For the detection of xa5, genotypes were screened using the primer xa5 s (multiplex). The primer amplified a 160 bp resistance allele in the check varieties IRBB59 and IRBB60 (Figure 2). On the other hand the susceptible check IR24 exhibited the presence of a 310 bp susceptible allele. In addition to the resistant checks (IRBB59 and IRBB60) the 160 bp allele contributing resistance governed by the xa5 gene was also detected in the genotype Bj1. Evaluation of the indigenous landraces revealed the presence of a 310 bp susceptible allele, indicating the absence of resistance attributed to the xa5 gene. The marker xa5 s (multiplex) has been earlier used for identifying resistant lines carrying 160 bp resistance allele for the gene xa5, as discussed in the works of Pradhan.
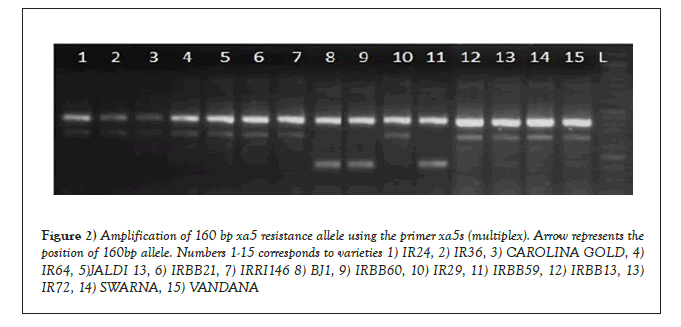
Figure 2: Amplification of 160 bp xa5 resistance allele using the primer xa5s (multiplex). Arrow represents the position of 160bp allele. Numbers 1-15 corresponds to varieties 1) IR24, 2) IR36, 3) CAROLINA GOLD, 4)IR64, 5)JALDI 13, 6) IRBB21, 7) IRRI146 8) BJ1, 9) IRBB60, 10) IR29, 11) IRBB59, 12) IRBB13, 13) IR72, 14) SWARNA, 15) VANDANA
The presence of a resistance conferring recessive allele for xa13 gene was confirmed using the primer xa13 Prom. The primer amplified a 540 bp resistance allele in the check varieties IRBB60 and IRBB59, along with the NIL IRBB13 (control for the xa13 gene). The susceptible check IR24 on the other hand amplified a 280 bp susceptible allele for the gene. In congruence with IRBB60, IRBB59 and IRBB13 a single genotype Bj1 exhibited the presence of a 540 bp resistance allele (Figure 3). The indigenous landraces on the other hand exhibited the presence of a 280 bp susceptible allele, which is similar to the susceptible check IR24. The primer xa13 Prom was successfully used in the earlier works of Magar [15-17].
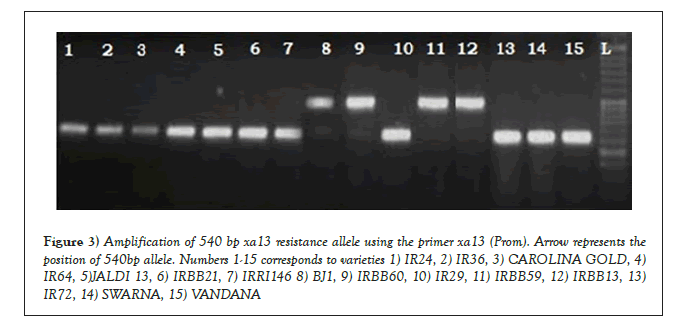
Figure 3: Amplification of 540 bp xa13 resistance allele using the primer xa13 (Prom). Arrow represents the position of 540bp allele. Numbers 1-15 corresponds to varieties 1) IR24, 2) IR36, 3) CAROLINA GOLD, 4)IR64, 5)JALDI 13, 6) IRBB21, 7) IRRI146 8) BJ1, 9) IRBB60, 10) IR29, 11) IRBB59, 12) IRBB13, 13) IR72, 14) SWARNA, 15) VANDANA
The dominant Xa21 was identified among the genotypes using the marker pTA248. The resistant checks IRBB60 and IRBB59 (carrying the dominant Xa21) along with the NIL IRBB21 (control for the Xa21 gene) amplified a 950 bp resistance allele (Figure 4). The primer pTA248 has been successfully used for identifying the 950 bp resistance allele during the investigations conducted by and Anjali [18]. The evaluation of the susceptible check IR24 revealed a 650 bp susceptible allele for the gene Xa21, when tested using pTA248. Apart from IRBB59, IRBB60 and IRBB21 no other genotype carried the dominant 950 bp allele for the gene Xa21; indicating the absence of resistance conferred by the gene. The cultivar Carolina Gold along with the landraces Kalabhat and Dudheswar exhibited the presence of a 750 bp allele for the gene Xa21 (Figures 5a-5d). The 750 bp allele has been designated earlier as a susceptible allele for the gene, as discussed in the works of Hittalmani [19].
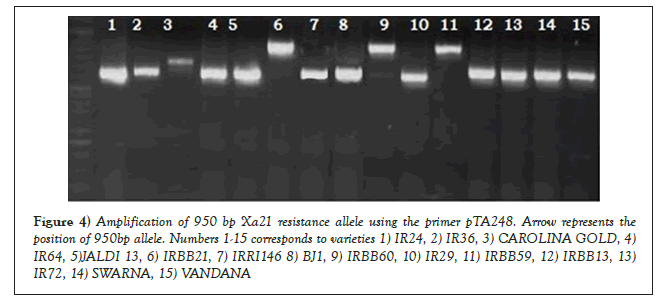
Figure 4: Amplification of 950 bp Xa21 resistance allele using the primer pTA248. Arrow represents the position of 950bp allele. Numbers 1-15 corresponds to varieties 1) IR24, 2) IR36, 3) CAROLINA GOLD, 4)IR64, 5)JALDI 13, 6) IRBB21, 7) IRRI146 8) BJ1, 9) IRBB60, 10) IR29, 11) IRBB59, 12) IRBB13, 13) IR72, 14) SWARNA, 15) VANDANA
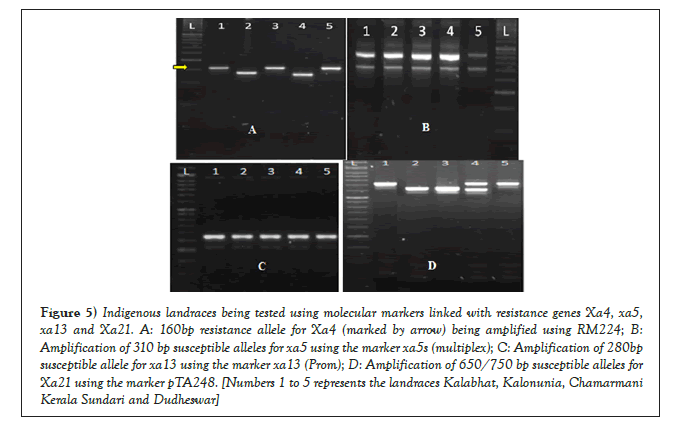
Figure 5: Indigenous landraces being tested using molecular markers linked with resistance genes Xa4, xa5, xa13 and Xa21. A: 160bp resistance allele for Xa4 (marked by arrow) being amplified using RM224; B: Amplification of 310 bp susceptible alleles for xa5 using the marker xa5s (multiplex); C: Amplification of 280bp susceptible allele for xa13 using the marker xa13 (Prom); D: Amplification of 650/750 bp susceptible alleles for Xa21 using the marker pTA248. [Numbers 1 to 5 represents the landraces Kalabhat, Kalonunia, Chamarmani Kerala Sundari and Dudheswar]
Phylogenetic analysis for determining genetic diversity among the genotypes
The cluster analysis classified the genotypes into four groups, which were designated as clusters I to IV (Table 5) (Figure 6). Cluster I was observed to be the largest, comprising of fifteen out of twenty genotypes analyzed in the current study. The genotypes representing cluster I exhibited variability in terms of resistance against the three virulent strains of X. oryzae. For the Xoo strains IX021 and IX027 the genotypes belonging to cluster I exhibited moderate resistance to complete susceptibility, whereas disease response to Xoo strain CUXo1 ranged from strong resistance (in case of Kalabhat and Chamarmani) to complete susceptibility (in case of Swarna, Vandana, JALDI 13, etc). Thus, under field conditions the genotypes representing cluster I can be expected to show variability in terms of pathogenesis, depending upon the strain infecting the cultivars.
| Cluster number | Genotypes | |||
|---|---|---|---|---|
| Range of pathogenicity for the strains | ||||
| IX021 | IX027 | CUXo1 | ||
| I | IR36, IR24, Vandana, Kerala Sundari, Jaldi 13, IR72, Kalonunia, Dudheswar, Kalabhat, Chamarmani,IR29,IRRI146, Swarna, IR64, IRBB13 | MR-SC | MR-SC | R-SC |
| II | BJ1 | SC | R | SC |
| III | IRBB21, Carolina Gold | R-MR | MS-MR | MR |
| IV | IRBB59, IRBB60 | R | R | R |
Note: MR= Moderately Resistant, MS= Moderately Susceptible, SC= Susceptible, R=Resistant
Table 5 Fifteen genotypes classified into four clusters along with the range of pathogenicity recorded for the clusters in response to the three strains of X. oryzae
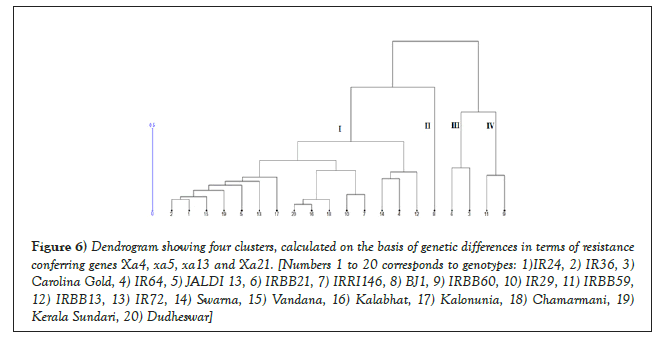
Figure 6: Dendrogram showing four clusters, calculated on the basis of genetic differences in terms of resistance conferring genes Xa4, xa5, xa13 and Xa21. [Numbers 1 to 20 corresponds to genotypes: 1)IR24, 2) IR36, 3) Carolina Gold, 4) IR64, 5) JALDI 13, 6) IRBB21, 7) IRRI146, 8) BJ1, 9) IRBB60 , 10) IR29, 11) IRBB59 , 12) IRBB13, 13) IR72, 14) Swarna, 15) Vandana, 16) Kalabhat, 17) Kalonunia, 18) Chamarmani, 19) Kerala Sundari, 20) Dudheswar]
The cluster II was represented by a single outlier Bj1, whereas cluster III was comprised of two genotypes, namely IRBB21 and Carolina Gold. The two resistant checks IRBB59 and IRBB60 were grouped together under cluster IV. The two genotypes exhibited stability in resistance against the three pathogenic strains. The cluster analysis further revealed that, cluster I being furthest from cluster IV, genetic distance between the two groups can be expected to be maximum. Thus popular cultivars like IR64, Jaldi-13, Swarna etc. belonging to cluster I can be hybridized with the resistant checks IRBB59 and IRBB60, which are grouped under cluster IV. Recombinant inbred lines generated from such crosses can be expected to show maximum genetic diversity in terms of resistance to BLB. Such diversity can be exploited in future breeding programs for introgression of desirable resistance genes from IRBB59 and IRBB60 into popular varieties cultivated in the region. Overall, the current study revealed that the three gene combination, i.e. xa5+xa13+Xa21 provided sufficient degree of stable resistance against the three X. oryzae strains. The contribution of the Xa4 gene was observed to be non-significant, especially when the remaining three resistance genes were present; as indicated by the similar degree of pathogenesis recorded for IRBB59 and IRBB60. The resistant check IRBB60 possessing the extra Xa4 gene (i.e. Xa4+ xa5+ xa13+ Xa21) did not show significant variation in pathogenesis when compared to the check variety IRBB59, which carried the three gene combination (i.e. xa5+xa13+Xa21). The weak response of Xa4 was further validated from the disease response observed in the genotypes IR29, IR36, IR64, JALDI 13 and Dudheswar. The genotypes carrying a single Xa4 gene could only manage to exhibit a moderate resistance in few cases with moderate to complete susceptibility in most of the cases, when inoculated with the three Xoo strains (Table 6). Slight exception to the phenomenon was recorded for the indigenous landraces Kalabhat and Chamarmani, which showed considerable resistance against the local Xoo strain CUX01. This can be partly attributed to the adaptive advantage of the landraces towards the local Xoo isolates. Consequently a slightly weaker response for the two landraces against the strains IX021 and IX027 revealed that the landraces are still vulnerable to BLB infections by new pathogenic strains. The weak response of Xa4 gene has been earlier reported in the works of French [20].
| Phenotypic response against the three strains |
Genotypes | Presence or absence of resistant alleles for the genes | Phenotypic response against the three strains | |||||
|---|---|---|---|---|---|---|---|---|
| Xa4 | xa5 | xa13 | Xa21 | IX021 | IX027 | CUX01 | ||
| 1 | IR36 | 1 | 0 | 0 | 0 | S | S | S |
| 2 | IR24 | 0 | 0 | 0 | 0 | S | S | S |
| 3 | IR64 | 1 | 0 | 0 | 0 | MR | S | S |
| 4 | Carolina Gold | 0 | 0 | 0 | 0 | MR | MS | MR |
| 5 | JALDI 13 | 1 | 0 | 0 | 0 | S | MS | S |
| 6 | IRBB21 | 0 | 0 | 0 | 1 | R | MR | MR |
| 7 | IRRI146 | 0 | 0 | 0 | 0 | MS | MS | S |
| 8 | IRBB60 | 1 | 1 | 1 | 1 | R | R | R |
| 9 | BJ1 | 0 | 1 | 1 | 0 | MR | R | MR |
| 10 | IR29 | 1 | 0 | 0 | 0 | MS | MS | S |
| 11 | IRBB59 | 0 | 1 | 1 | 1 | R | R | R |
| 12 | IRBB13 | 0 | 0 | 1 | 0 | MR | S | MS |
| 13 | IR72 | 0 | 0 | 0 | 0 | S | MS | MS |
| 14 | Swarna | 0 | 0 | 0 | 0 | MR | MS | S |
| 15 | Vandana | 0 | 0 | 0 | 0 | S | S | S |
| 16 | Kalabhat | 1 | 0 | 0 | 0 | MR | MS | R |
| 17 | Kalonunia | 0 | 0 | 0 | 0 | S | S | S |
| 18 | Chamarmani | 1 | 0 | 0 | 0 | MR | MS | R |
| 19 | Kerala Sundari | 0 | 0 | 0 | 0 | S | S | S |
| 20 | Dudheswar | 1 | 0 | 0 | 0 | MS | MS | MS |
Note: 1= presence of resistance allele, 0=absence of resistance allele
Table 6 Presence or absence of resistance allele for the genes Xa4, xa5, xa13 and Xa21 along with the degree of pathogenesis observed in twenty genotypes inoculated with three pathogenic strains of X. oryzae
The current study also revealed that the presence of a single xa13 gene did not provide sufficient protection against the three X. oryzae strains [21]. The near isogeninc line IBBB13 possessing a single xa13 gene showed moderate resistance to complete susceptibility against the three pathogenic strains. Contrary to xa13, the dominant Xa21 when present singly conferred significantly better protection against the pathogenic strains, as observed in the case of IRBB21 [22]. The disease response for IRBB21 revealed that the gene Xa21 when present singly, could manage to confer moderate to complete resistance against the three pathogenic strains [23]. Thus, the introgression of dominant Xa21 gene into commercially popular cultivars may prove to be crucial for ensuring a durable resistance against multiple pathogenic strains of X. oryzae (Figures 7 and 8). The relation between various resistance conferring gene combinations and the degree of pathogenesis recorded among the genotypes are further illustrated in Figures 7 and 8.

Figure 7: (A) and (B) represents images of infected leaves of IRBB13 and IRBB21 exhibiting complete susceptibility and moderate susceptibility respectively against the strain IX027. (C) represents image of the infected leaf of IRBB60 infected by IX027.
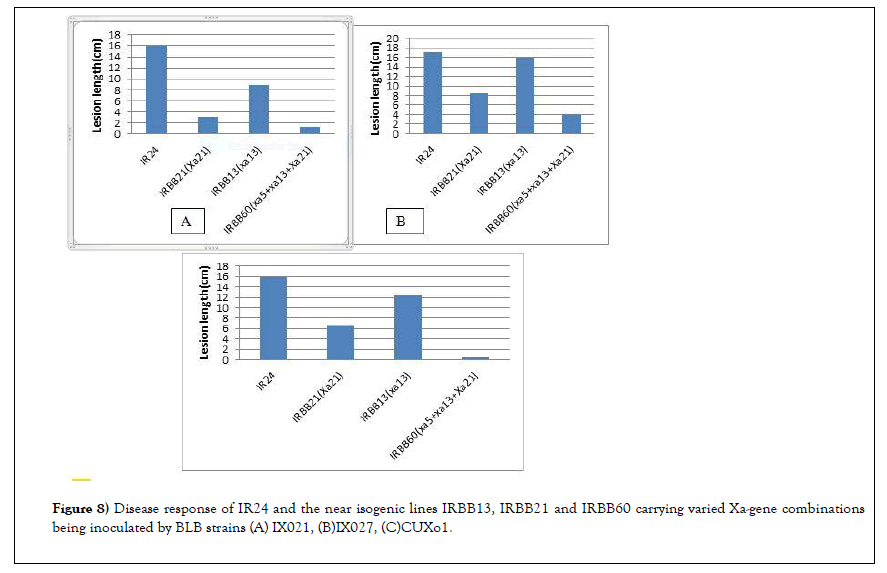
Figure 8: Disease response of IR24 and the near isogenic lines IRBB13, IRBB21 and IRBB60 carrying varied Xa-gene combinations being inoculated by BLB strains (A) IX021, (B)IX027, (C)CUXo1.
The current study successfully evaluated the range of pathogenesis observable among rice genotypes, in response to the infection caused by three virulent strains of X. oryzae. Based on the phenotypic and genotypic screening of the cultivars it was evident that the three resistance gene combination i.e. xa5+xa13+Xa21 was vital for establishing a steady resistance against the pathogenic strains used in the current investigation. On the other hand lines possessing a single or two resistance conferring genes showed variable pathogenesis when infected with the three X. oryzae strains. The investigation further revealed that popular cultivars in West Bengal, namely Swarna, IR64, Vandana, as well as the indigenous landraces of the region like Dudheswar, Kerala Sundari and Kalonunia are particularly vulnerable to X. oryzae infection. The investigation also identified the presence of dominant Xa4 gene among the indigenous landraces Kalabhat and Chamarmani, which showed strong resistance against the local Xoo strain CUXo1. However, the two landraces failed to show significant resistance against the strains IX021 and IX027, indicating that the landraces are still vulnerable to infections resulting from newly introduced Xoo strains in the region. Finally, the cluster analysis revealed that the popularly cultivated varieties in West Bengal, namely Swarna, IR64, Vandana, as well as commercially desirable landraces like Chamarmani and Dudheswar exhibited significant allelic variation in respect to BLB resistance genes from the check varieties IRBB59 and IRBB60. Thus hybridization between the popular cultivars and the resistant checks can ensure significant diversity among recombinant inbred lines. This diversity can be exploited in later breeding programs, designed for developing BLB resistant cultivars suited for West Bengal as well as rest of India.
The authors extend their gratitude towards the project “Crop Breeding Research Unit” funded by Department of Agriculture, Govt. of West Bengal, India and Rice Research Station, Chinsurah, Hooghly Govt. of West Bengal for providing infrastructural support.
Citation: Nath D, Nath A, Majumder K, et al. Phenotypic and molecular screening of rice genotypes inoculated with three widely available pathogenic strains of Xanthomonas oryzae pv. oryzae. AGBIR. 2022; 38(2):278-284.
Received: 03-Mar-2022, Manuscript No. AGBIR-22-56650; , Pre QC No. AGBIR-22-56650 (PQ); Editor assigned: 07-Mar-2022, Pre QC No. AGBIR-22-56650 (PQ); Reviewed: 21-Mar-2022, QC No. AGBIR-22-56650; Revised: 28-Mar-2022, Manuscript No. AGBIR-22-56650 (R); Published: 04-Apr-2022, DOI: 10.35248/0970-1907.22.38.278-284
Copyright: This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http:// creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
