Agricultural and Biological Research
RNI # 24/103/2012-R1
Research - (2022) Volume 38, Issue 6
Bacillus thuringiensis (Bt.), gram-positive, spore-forming bacteria, is pervasive and can be found in all kinds of natural environments. Several methods have been used to isolate and characterize Bt. for this investigation. Fourteen Bt. in North Gondar, Ethiopia, isolates were found in eight different soils. Bt. the physical properties of the colonies, the structure of the parasporal crystals, and the PCR-amplification profiles were used for identifying. For both sets of primers, including those for the cry1 and cry9 genes, 12 strains demonstrated successful results. 50 Bt. strains were found among the 72 bacterial strains that were examined to have 4 different forms of crystalline inclusions. The majority of the 15 isolates (30%) had irregular pointed Bt. crystals, while 20, 24, and 26% of the strains had bi-pyramidal, cuboidal, and spherical crystals, respectively. According to the PCR results, 50% of isolates amplifying the cry 9 gene and the cry1 gene (35.71%), indicating that the cry 9 gene frequency predominated. Two strains did not amplify. Bt. variety was evaluated using a combination examination of their protein and PCR banding patterns. This study reveals that the distribution of Cry-type genes can be used to classify and identify Bt. isolates that were recovered from soil samples in the North Gondar Zone.
Bacillus thuringiensis; Cry gene; PCR; Primer
A gram-positive, rod-shaped bacterium called Bacillus thuringiensis (Bt.) generates endotoxin, a crystal protein that is insecticidal [1]. During sporulation, these bacteria produce one or two crystalline parasporal inclusions, Cry and Cyt, which are encoded by the respective cry and cyt genes [2]. The lepidopteron, dipteran, and coleopteran orders of insect pests are therefore among those that Bt. has the most ability to manage [3]. Recent research [4,5] on Bt's toxicity against several insect pests has been published. The toxicity of Bt. against many insect pests and disease vectors has been the subject of numerous academic articles [4]. Three classes of Cyt proteins (Cyt1-Cyt3) and 74 classes of Cry proteins (Cry1 to Cry74) have been demonstrated to have distinct binding to the receptor based on their amino acid sequence homology [6]. These crystal proteins are protoxins that are proteolytically broken down into smaller poisonous polypeptides in the midguts of insects. Active poisons cause target species' epithelial cell membranes to rupture through osmotic lysis [7]. These cry proteins have no effect on people or other animals. The increased resistance of insect pests leads to the usage of synthetic chemical pesticides [8]. Therefore, continuing research is being done to find novel Bt. strains with a particular host repertoire or a higher potential for toxicity. Industrial Bt-based biopesticides are still employed at 10–50 g/acre (1020 molecules/acre) globally, whereas organophosphates and pyrethroids are utilized at 8: 1024 and 3: 1022 molecules/ac, respectively, of conventional chemical pesticides [9]. Thus, the molecular size of these toxins is around 80,000 times greater than that of organophosphates and 300 times greater than that of synthetic pyroids [10]. Studying new Bt. strains may lead to the identification of novel insecticidal proteins that are more toxic, which will be essential in providing alternatives to deal with the advent of resistant insect populations. Multiple novel Bt. strains must therefore be isolated from various environmental settings. There are numerous novel techniques for cloning insecticidal crystal protein genes [10]. The polymerase chain reaction (PCR) has been widely used to classify Bt. strains [11]. The simultaneous sampling of many Bt. strains and this method's high sensitivity to the rapid detection and recognition of genetic sequences aid in the identification and forecasting of pest behavior [12]. One of the best places to find genetic variety and biodiversity is Ethiopia. The scientific publications of Bt. diversity from Ethiopia, however, lack information [13]. As a result, the goal of this work was to identify, classify, and bioassay a number of Bt. strains expressing the novel cry gene from several sites in North Gondar Zone, Ethiopia. Future insect biocontrol initiatives may benefit from the current study's findings.
Sample collection
The study area was in Ethiopia's Gondar Zone, in the Amhara region. In northwest Ethiopia, samples of the soil were taken from two different locations. Using a sterile spatula, all of the soil samples (8 g each) were collected from a depth of 2 to 5 cm. prior to usage; the samples were kept in storage at 4°C.
Reference strain
Bacillus thuringiensis subsp. kurstaki HD1 was made available by the Ethiopian Biodiversity Center (Addis Abeba, Ethiopia) as a positive control.
Isolation of strains
Sodium acetate was used in the study to prevent the germination of spore-bearing bacteria present in the samples and heat treatment was used to kill non-spore-bearing bacteria. Each sample was precisely weighed at one gram, added to tubes containing 10 ml of liquid Luria and nutrient broth medium, thoroughly mixed with 0.25 molar sodium acetate, and then incubated for four hours at 30°C at 200 rpm. The test tubes were then heated for three minutes at 8°C by adding the hot water base. Following that, the solution was serially diluted with sterile water up to a multiplicity of 10-5 (10-1, 10-2, 10-5) 20 microliters of the supernatant from each dilution sample (10-1, 10-2, 10-5)were aseptically distributed on nutritional agar plates and incubated at 30°C for 24 hour.
Bt. isolation using the selective medium
Except for Bt, L-Serine can inhibit the growth of Bacillus species. M9 minimal medium was employed as a selective medium and included 0.2 mM of L-Serine per liter [14]. On nutritional agar medium containing 0.25 M sodium acetate, bacterial colonies were produced. On a selective medium of L-serine, the colonies grown on nutrient agar medium were cultivated for 48 hours. The next phase was selecting a white single colony and examining the strains for protein crystals. Culture smears were produced, heat-fixed, and stained with a 0.133% in 50% acetic acid Coomassie Brilliant Blue G250 stain solution. The smears were then carefully cleaned under running tap water, dried off, and bright-field analyzed to check for crystalline inclusions. Based on the presence of crystalline inclusions, Bt. strains were chosen.
Molecular identification of cry gene screening in indigenous Bt. strains
For cry gene screening, two sets of universal primers were utilized in PCR with Bt. genomic DNA. Using a DNA extraction kit, we extracted DNA in accordance with the manufacturer's instructions for gram-positive bacteria (HIMEDIATM Laboratories Pvt. Ltd Company, India). Cry1 and Cry9 primers were used in PCR reactions (manufactured by Eurofins Genomics India Company) (Table 1). The PCR for the Cry genes was carried out in a total volume of 20 μl, which included 5 μl of 1X PCR buffer (10 mM Tris-HCl, pH 8.0 at 25°C, 1.5 mM MgCl2), 2 μl of bacterial genomic DNA, 20 pM/μl each of the forward and reverse primers, 0.4 μl of Taq DNA polymerase, 1 μl of 200 mMd NTPs, 1.5 μl of 3 mM MgCl2, and 9 μl of sterile double distilled water in a final volume of 20 μL. Using a step-cycle program package, amplification was carried out on a DNA thermal cycler for 35 cycles, with each cycle consisting of denaturation for two minutes at 94°C, one minute at 95°C, annealing for one minute at 48°C for cry1, elongation for five minutes at 72°C, and final extension for ten minutes at 72°C for cry1. For the cry9 genes, 35 amplification cycles that included denaturing at 94°C for 1 minute, annealing at 600°C for 1 minute, elongating at 72°C for 1 minute, and finally extending at 72°C for 10 minutes were performed [15]. After amplification, 10 µl of each PCR product was electrophoresed in 0.8% -1.5% agarose-EtBr gel (Tris-Borate EDTA electrophoreses, pH 8.0) at 100 V for 40 min. The DNA bands were then seen using a gel documentation system [16].
| Genes selected | Sequence (5’-3’) | Product size (bp) | Annealing temp. (°C) |
|---|---|---|---|
| Cry1 | TGTAGAAGAGGAAGTCTATCCA | 272-290 | 48 |
| TATCGGTTTCTGGGAAGTA | |||
| Cry9 | CGGTGTTACTATTAGCGAGGGCGG | 351-354 | 72 |
| GTTTGAGCCGCTTCACAGCAATCC |
Table 1: Primer sequences for different cry genes.
Statistical data analysis
The proportions of Bt. isolates from various sample sources, locales, Bt. indices, and molecular patterns of Cry genes were documented in the data [17,18].
Characterization of Bt. morphology
Bt. isolates have been discovered in a wide range of ecosystems, including soil microflora and marine habitats [19,20]. Bt. was employed in 32% of the 503 soil samples collected from around Ethiopia's 16 agro-ecological zones [13]. For this study, soil samples from several places in North Gondar, Ethiopia, were used to extract 16 Bt. strains. Similar findings were reported by a number of researchers. [9] 159 soil samples were taken from the Kashmir valley in the Himalayas, and a total of 68 Bt. strains were isolated. Similar to this, [1] identified 317 Bt. strains from 231 samples gathered throughout the 26 districts of Bangladesh, encompassing six different regions. The most common method for categorizing Bt. is to search for crystalline inclusions. For the high-throughput examination of stained bacterial colonies for the presence of crystals and the detection of small crystals, microscopy is more useful than phase-contrast microscopy [21]. Both of the Bt. isolates tested positive for gram-positive bacteria and formed endospores. Similar to this, several crystal morphologies were noted. The form of bipyramidal crystals was most frequently observed, followed by spherical, irregularly pointed, and cubical crystal types. The 14 Bt. strains had different crystalline inclusion shapes (Figure 1). Identified Bt. isolates based on hemolytic behavior and the presence of parasporal crystal proteins in a related discovery. The isolates formed irregular white colonies with a pink background and included four different types of parasporal crystal proteins, including bipyramidal, spherical, irregularly pointed and cuboidal shaped crystal proteins. This demonstrates the diversity of the local Bt. isolates [20]. Isolated Bt-like colonies were identified using Gram, spore, and crystal stains. 21 of the 50 bacterial isolates included crystalline inclusions, which were identified as Bt. by bright-field microscopy. Bt. indices of 0.69 allowed the isolation of 12 Bt. strains from soil samples collected in North Gondar, Ethiopia (Table 2). The 0.86 Bt. indices in their samples from Bangladesh were noted in earlier research [1]. Recent research have published various Bt. the index values, with values in India ranging from 0.028 to 0.075 [9]. According to, both biotic and abiotic environmental factors, including nutrient availability, texture, pH, temperature, and humidity, may have an impact on the Bt. index [22]. Biotic environmental factors include the vegetal top, the type of insect that is typically found in the area, or soil microorganisms. Because of these factors, they refrain from comparing their findings to those of other researchers. In our research, we noticed some variations in the frequency of Bt. isolation from a single sample set. However, it is impossible to pinpoint the precise elements that affected the dispersion in various ecosystems. Therefore, additional research into the relationship between the quantity and dispersion of Bt. and the Physico-chemical characteristics of the habitats of the study locations may be necessary.
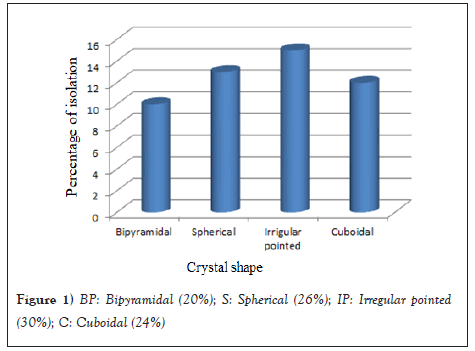
Figure 1: BP: Bipyramidal (20%); S: Spherical (26%); IP: Irregular pointed (30%); C: Cuboidal (24%).
| Type of the sample source | Total number of Samples Collected | Total number of colonies obtained | Total number of isolates for Bt | Bt Index |
|---|---|---|---|---|
| Soil | 8 | 72 | 50 | 0.69* |
Note: * Bt. An index is the ratio of the total number of isolates for Bt. to the total number of colonies obtained from samples (soil).
Table 2: Bacillus thuringiensis isolated from different sample sources.
PCR-based screening of cry genes in native Bt. strains
By amplifying the estimated size of PCR products using two sets of universal primers, the presence of cry-type genes in Bt. strains was verified. The majority of the cry-type genes, led by cry9 (50%) in the local Bt. strains, were present in cry1 (35.71%) (Table 3). Likewise, among the evaluated Cry-type genes, Cry1 (83.33%) genes were the most common, followed by cry2 (38%), cry4 (27.77%), and cry3 (16.6%) genes [23]. The prevalence of genes displayed was based on the frequency reported by many researchers. According to [15], eight Bt. strains (IS1-IS8) included Cry-type genes. As a result, all strains of indigenous isolates contained cry1 type genes, which were found to be the most prevalent by PCR. These genes were followed by vip 3A (87.5 %), cry2 (75%), cry9 (62%), cry3 (50%), cry11 (37.5%), cry7-8 (37.5%), cry5, 12, 14, 21 (25%), cyt1 (25%), cry4 (12.5%), and cyt2 (12.5%). These results reveal the influence of geographic areas on the diversity of cry gene content in Bt. strains. The results show that Cry1 and Cry9 encode Lepidoptera-specific toxins. 12 isolates that possess cry1 and cry9 are hence at least hypothetically toxic to Lepidoptera.
| S.No. | Isolate code | Cry gene | Sample type | Site of the sample collected | % of isolates tested by PCR |
|---|---|---|---|---|---|
| 1 | LK-1 | Dembia | 35.71% | ||
| 2 | LM-11 | Gondar zuria | |||
| 3 | LM-8 | 1 | Soil | Gondar zuria | |
| 4 | LM-9 | Dembia | |||
| 5 | LK-5 | Dembia | |||
| 6 | LM-4 | Dembia | |||
| 7 | LM-7 | Dembia | |||
| 8 | LK-12 | Gondar zuria | 50% | ||
| 9 | LM-3 | 9 | Soil | Dembia | |
| 10 | LM-2 | Gondar zuria | |||
| 11 | LM-10 | Dembia | |||
| 12 | LM-14 | Dembia and | |||
| 13 | LK-6 | 12 | Gondar zuria | Did not react with two primer | |
| 14 | LK-13 | Gondar zuria | |||
| Total | 14 | 85.71% |
Table 3: Annual total amount of yam plant in ha damaged by elephants and estimated monetary value.
On the basis of amino acid sequence homology, it has also been demonstrated that three cyt protein classes (Cyt-Cyt3) and 74 cry protein classes (Cry1 to Cry74) exhibit distinct binding to the receptor [6]. Previous research found that different geographical areas of the world had varying percentages of isolates that did not produce PCR products (between 14% and 40%) [23]. All of these discoveries suggest that unique and unexplained crystalline protein genes are present in many isolates worldwide. The results showed that a systemic, extensive sampling of Bt. isolates could be carried out using a PCR-based method in order to identify cry genes and assess their toxicity. By amplifying the PCR results to their predicted sizes in the two primers, the presence of the aforementioned cry-type genes and the distribution of Bt. strains were verified. As a result, the use of PCR has greatly improved cry gene identification. However, this method is limited to individuals belonging to already known gene families and calls for the use of several primers.
Morphological and molecular characterization methods, such as crystalline inclusions and targeted PCR of Cry-type genes, can be used to discriminate and identify Bt. Due to this genetic variety among isolates, even those from the same sample showed significant variance in terms of their crystalline protein composition and insecticidal action due to variations in their cry-type genes. The location of the samples may be a factor in the difference and activity. Out of the 14 isolates, 12 amplify using the two tested pairs of primers. Two isolates failed to amplify any of the study's primers. The findings of this study support the necessity for ongoing research into new Bt. strains from various global environments. The field of integrated pest management will benefit from further research on gene transfer and the characterization of novel cry genes from these isolates of Bt.
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Tarekegn M, Salillih F. Molecular Characterization of Bacillus thuringiensis Strains from Soil Sample in North Gondar, Ethiopia. AGBIR.2022; 38(6):392-394.
Received: 02-Nov-2022, Manuscript No. AGBIR-22-78890; , Pre QC No. AGBIR-22-78890 (PQ); Editor assigned: 07-Nov-2022, Pre QC No. AGBIR-22-78890 (PQ); Reviewed: 21-Nov-2022, QC No. AGBIR-22-78890; Revised: 25-Nov-2022, Manuscript No. AGBIR-22-78890 (R); Published: 02-Dec-2022, DOI: 10.35248/0970-1907.22.38.392-394
Copyright: This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http:// creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
